Translate this page into:
Investigating the presence and virulence potential of Enterococcus faecalis, with a focus on the hyaluronidase factor, in environmental samples: Insights from a pilot study
*Corresponding author: C. Benedict Paul, Department of Biotechnology, Sri Ramachandra Institute of Higher Education and Research, Chennai, Tamil Nadu, India. benedictpaulc@sriramachandra.edu.in
-
Received: ,
Accepted: ,
How to cite this article: Dhanushiya R, Abinaya S, Siva V, Paul CB. Investigating the presence and virulence potential of Enterococcus faecalis, with a focus on the hyaluronidase factor, in environmental samples: Insights from a pilot study. Sri Ramachandra J Health Sci. 2024;4:53-8. doi: 10.25259/SRJHS_55_2023
Abstract
Objectives:
Enterococcus faecalis is implicated in various human diseases, including urinary tract infections, dental conditions such as periodontitis, dental caries, systemic disorders such as infective endocarditis, meningitis, intra-abdominal infections, and wound infections. The virulence genes associated with these diseases include adhesins (ace, efaA, efaB, ebp), biofilm-forming factors (esp, gelE, fsr), cytolysin (cylA, cylB, cylM, cylL), hyaluronidase (hyl), aggregation substance (asp), gelatinase (gelE), enterococcal surface protein (esp), among others. In a prior investigation, we observed a higher prevalence of the hyl gene in medical isolates, prompting an exploration of its occurrence in the environment.
Material and Methods:
32 samples from diverse locations in and around Chennai were collected. Enterococcus spp. isolation was achieved using a partially selective medium (Ethyl Violet Azide medium). Subsequently, the isolates were identified as E. faecalis through polymerase chain reaction targeting the 16S ribosomal RNA (rRNA) gene. Positive 16S rRNA samples were further scrutinized for the presence of the hyl gene using Sequence specific oligonucleotide primers.
Results:
Our findings indicated an E. faecalis incidence of 41%. However, none of the isolates tested positive for the hyl virulence gene.
Conclusion:
This study suggests that the hyl gene may play a pivotal role in pathogenesis, as previous investigations in our laboratory involving medical isolates reported a 71% incidence of the hyl gene. In addition, our results highlight the opportunistic nature of the organism.
Keywords
Enterococcus faecalis
Virulence genes
Environmental prevalence
16S ribosomal ribonucleic acid gene
Opportunistic pathogen
INTRODUCTION
Enterococcus faecalis, a Gram-positive bacterium, is a ubiquitous inhabitant of various ecological niches, including the gastrointestinal tracts of both humans and animals, as well as soil, water, and food sources. While typically regarded as a commensal microorganism, E. faecalis can transition into an opportunistic pathogen by acquiring specific virulence genes. This bacterial species has been identified as a significant contributor to nosocomial infections, constituting approximately 90% of all enterococcal infections in humans, with a notable predilection for individuals with compromised immune systems.[1,2] The genomic landscape of E. faecalis, encompassing a circular genome of 3 million base pairs and 3000 genes, provides a comprehensive understanding of its biological intricacies and evolutionary pathways. Notably, E. faecalis has developed intricate mechanisms to counteract antibiotics, including the alteration of targets, modification of enzymes, and even autonomous antibiotic production. These mechanisms collectively contribute to the resilience of E. faecalis strains against conventional antibiotic treatments, posing challenges in the clinical management of infections caused by this bacterium.[3] At the heart of E. faecalis’ pathogenicity lies an array of virulence factors, each playing a crucial role in host colonization and the establishment of infection. One such factor of particular interest is hyaluronidase (Hyl), encoded by the hyl gene. Hyl is a degradative enzyme known for its involvement in tissue damage. Functionally, it depolymerizes the mucopolysaccharide components of connective tissues, enhancing the invasiveness of bacteria. This enzyme, identified in various organisms, acts as a potent spreading factor, facilitating the movement of bacteria and their toxins through host tissues.[4] Hyl activity facilitates the spread of infection by breaking the extracellular matrix potentially impacting the stability and resistance that influence the severity and persistence of oral related disease. The detection of the hyl gene in oral microbiota can serve as a biomarker for the presence of pathogenic bacteria, which can be useful in diagnosing oral infections. The focus of the present research extends beyond clinical settings to explore the prevalence of E. faecalis in diverse environmental samples, including soil, water, bird feces, and animal feces in the vicinity of Chennai. Furthermore, this study seeks to unravel the distribution and implications of the hyl gene in environmental isolates of E. faecalis. By examining these factors in a broader context, the research aims to contribute valuable insights into the ecological dynamics and potential public health ramifications associated with this pathogenic bacterium and its virulence factors in various environmental reservoirs.
MATERIAL AND METHODS
Ethics approval
The collection of environmental samples for this study was carried out in strict adherence to ethical guidelines. The Institutional Ethics Committee (IEC) granted the necessary approval for the conduct of the study (CSP/23/JUN/130/540).
Sample collection
A total of 32 environmental samples were procured from various locations in and around Chennai. The collection process employed sterile 15 mL Falcon tubes to ensure the integrity and uncontaminated nature of the samples. 10 mL of water sample was collected using sterile falcon tubes. For solid samples, 5 g was collected from different environments. As this is a pilot study, no specific criteria were applied for sampling except that the samples should be of different types such as water and soil. The details of the sample collection are given in Table 1.
| S. No. | Sample type | Location |
|---|---|---|
| S1 | Soil | Redhills |
| S2 | Soil | Guduvancheri |
| S3 | Bird feces | Kattupakkam |
| L4 | liquid | Redhills |
| S5 | Soil | Nungambakkam |
| S6 | Soil | Pattinapakkam |
| S7 | Soil | Aminjikarai |
| S8 | Soil | Lake |
| S9 | Soil | Park |
| S10 | Soil | Guduvanchery Bus Stand |
| S11 | Soil | Iyyappanthangal Depot |
| S12 | Soil | Anna Nagar East |
| L13 | Water | Aminjikarai |
| S14 | Soil | Mogappair |
| S15 | Soil | Sinthamani |
| S16 | Soil | Iyyappanthangal |
| S17 | Soil | Porur |
| L18 | Liquid | Porur |
| S19 | Soil | Karayanchavadi |
| S20 | Soil | Kumananchavadi |
| S21 | Soil | Poonamallee |
| L22 | Liquid | Mangadu |
| S23 | Soil | Lake2 |
| S24 | Soil | Pond Guduvancheri |
| L25 | Liquid | Guduvancheri Water |
| S26 | Soil | Guduvancheri Garden |
| S27 | Cow feces | Shenoy Nagar |
| S28 | Goat feces | Shenoy Nagar |
| S29 | Crow feces | Shenoy Nagar |
| S30 | Cow feces | Cuddalore |
| L31 | Cow shed | Cuddalore |
| S32 | Hen feces | Cuddalore |
Sample processing
To prepare the samples, 4.0 mL of distilled water was added to 1.0 g of solid samples in Falcon tubes. Subsequently, 2.5 mL of the resulting diluted sample was mixed with an equal volume of Ethyl Violet Azide broth (EVA broth). For water samples, 2.5 mL directly combined with 2.5 mL of EVA broth. The tubes were then subjected to a 48-h incubation period.
Bacteriological isolation and biochemical identification
Bacterial isolation was conducted using EVA agar, a partially selective medium specifically inhibiting the growth of nonenterococcal Gram-positive and Gram-negative bacteria due to the inclusion of sodium azide. After 24–48 h of incubation at 37°C, gray colonies indicative of enterococcal species were observed. These gray colonies were subcultured onto Brain Heart Infusion agar. In addition, colonies were streaked onto Bile esculin agar plates to confirm the presence of Enterococcus species through esculin hydrolysis, resulting in brownish-black precipitate colonies. All dehydrated culture media were procured from (HiMedia™ Laboratories Pvt Ltd).
DNA extraction
DNA extraction from all samples utilized the boiling lysis method as described by Dashti et al. and Yost and Nattress.[5,6] Following centrifugation of 1.0 mL of the broth culture for 5 min at 10,000 rpm, the supernatant was discarded, and 200 mL of sterile distilled water was added. The mixture was then subjected to a dry bath at 95°C. Two microliters of the resulting supernatant served as the template in polymerase chain reaction (PCR), as per the method outlined by Mullis et al.[7] Extracted DNA was stored at −20°C for subsequent analysis.
Molecular identification using 16S ribosomal RNA (rRNA)
Primer designing
Enterococcus isolates were further identified as E. faecalis using Sequence-Specific Oligonucleotide Primers (SSOPs) in PCR. Primers were designed with the assistance of the online primer designing tool, Primer3.[8] The 16S rRNA gene sequence, retrieved from the Nucleotide database of National Center for Biotechnology Information (accession id: AB036835.1), was submitted to the Primer3 tool. Default parameters were slightly modified to ensure the product size of the amplicon fell between 500 and 1000 nucleotides. The pick primer option was employed, and the results are illustrated in Figure 1. The melting temperatures of the forward and reverse primers were 59.03°C and 59.01°C, respectively.
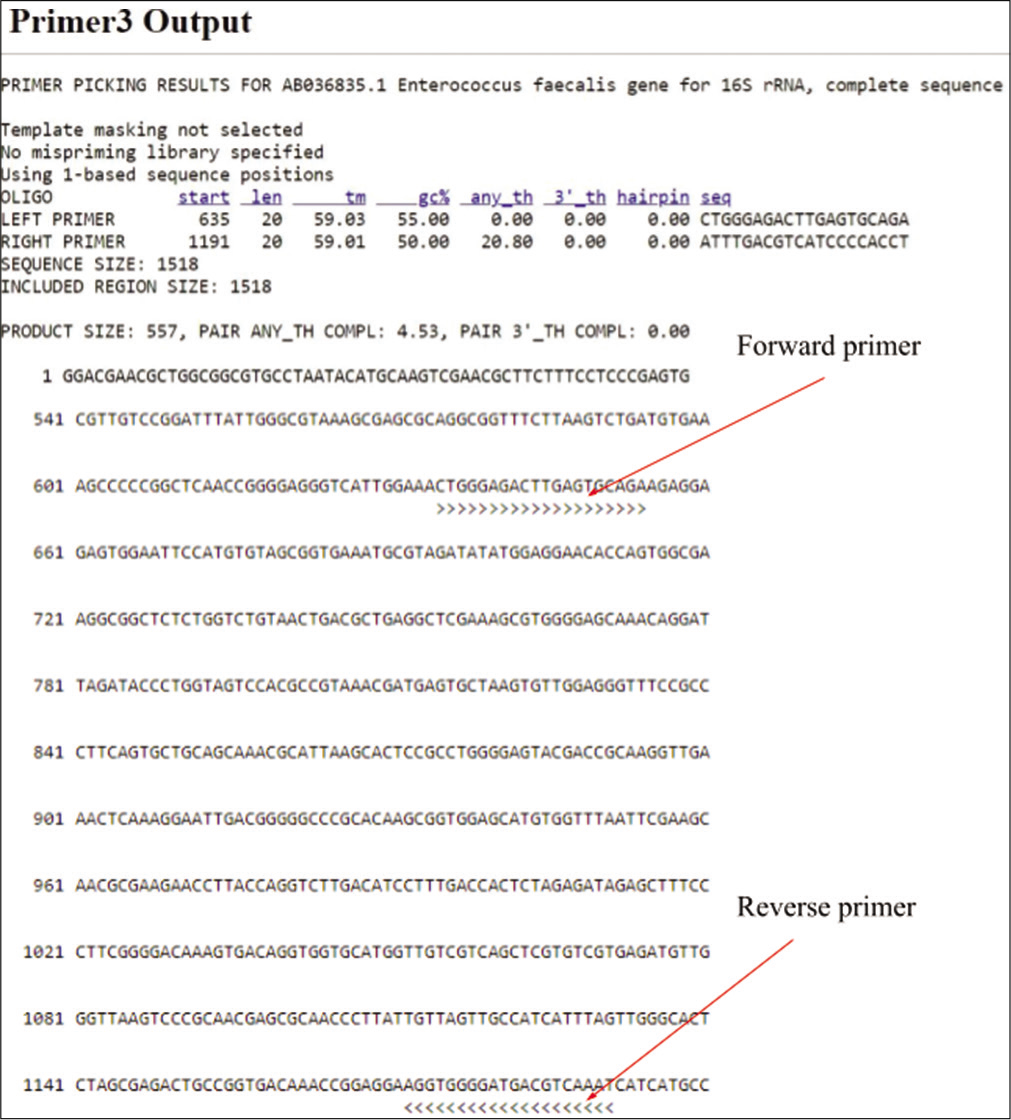
- Primer designing using primer 3.
Molecular detection
For the molecular identification of all isolates in the study, PCR amplification of the E. faecalis 16S rRNA gene was performed and the primers were as follows: Forward primer 5’CTGGGAGACTTGAGTGCAGA3’ and Reverse 5’ATTTGACGTCATCCCCACCT3’ (Sigma Aldrich, USA). The primer details are given in Table 2. A primer concentration of 0.4 mM was employed. PCR was executed using Taq DNA Polymerase 2x Master Mix RED (Ampliqon, Denmark) in a 20 µl reaction maintaining a pH of 8.5 with Tris-HCl pH 8.5, ammonium sulfate. It also has 3 mM magnesium chloride, 0.2% Tween®20, 0.4 mM of each deoxynucleotide triphosphate, and 0.2 units/µL Taq DNA polymerase. Moreover, the TaqMix is added with an inert red dye for tracking in the agarose gel. The cycling parameters for amplification were as follows: Initial denaturation - 95°C; 5’, denaturation - 95°C; 45”, annealing - 60°C; 45”, extension - 72°C; 1’, repeated for 30 cycles. Subsequent to an 8-min final extension at 72°C, the reaction mixtures were cooled to 4°C. Gel electrophoresis of the PCR products was carried out on 1.2% agarose gel and visualized using an ultraviolet (UV) Transilluminator (White/UV transilluminator UVP). The 2 Kbp DNA ladder (Thermo Fisher Scientific, US) was used in all agarose gel electrophoresis experiments.
| Primers | Nucleotide Sequence (5’–3’) | Expected product size | Reference |
|---|---|---|---|
| Forward | 5’–CTGGGAGACTTGAGTGCAGA–3’ | 557 bp | Designed in this study |
| Reverse | 5’–ATTTGACGTCATCCCCACCT–3’ |
16S rRNA: 16S Ribosomal RNA, bp: Base pairs, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
Detection of the virulence factor, hyl
hyl primers
The design of hyl primers followed the procedure outlined earlier for the 16S rRNA primers. Primers were designed and synthesized based on the sequences presented in Table 3. Subsequently, PCR was conducted utilizing these primers to amplify the region of interest associated with the hyl virulence factor.
| Primer (hyl) | Nucleotide Sequence (5’– 3’) | Expected product size | Reference |
|---|---|---|---|
| Forward | 5’CCCTGGACACATGAAATGCG 3’ | 501 bp | Designed in this study |
| Reverse | 5’AGCATCGGCCGTTGATAGAC 3’ |
Hyl: Hyaluronidase, bp: Base pairs, A: Adenine, C: Cytosine, G: Guanine, T: Thymine
PCR-cycling parameters
For the 16S rRNA PCR of the hyl gene, the cycling conditions are as follows: Initial denaturation - 95°C; 5’, denaturation - 95°C; 45’’, annealing - 60°C; 45”, extension - 72°C; 1’ with 30 cycles. Following an 8-min final extension at 72°C, the reaction mixtures were then cooled to 4°C.
Visualization and documentation
PCR products were run on a 1.2% agarose gel that has ethidium bromide at a working concentration of 0.2 mg/µL (16 µL/80 µL). Electrophoresis was conducted in a 1× TBE buffer for 30 min at 70V. Subsequently, bands were visualized under a UV transilluminator (UVP Upland, USA) and documented using a Gel Documentation System (Vilber Lourmat, USA). Molecular Marker 16S rRNA PCR was conducted for all isolates, with primers designed specifically for detection, yielding an amplicon size of 557bp. E. faecalis ATCC 29212 served as the positive control, and a 2 Kbp DNA ladder (Thermo Fisher Scientific, US) was used as a marker with 500 bp as the lowest band. DNA bands were observed in all wells except the negative control, indicating the absence of contamination. The results were documented using the Gel Documentation instrument and UV illuminator (White/UV Transilluminator UVP).
RESULTS
Isolation and biochemical identification
For the biochemical identification of Enterococcus species, EVA plates were employed to culture the isolates. Following an overnight incubation period, colony morphology on EVA plates were scrutinized. The isolates inoculated onto EVA plates and incubated at 37°C showed the formation of typical opaque, entire, and pinpoint colonies while reading. Subsequent subculturing of the 32 isolates on EVA agar plates revealed 30 isolates showing grayish pinpointed colonies characteristic of E. faecalis. Notably, isolates S16 and L4 were exceptions, yielding negative results in this biochemical identification process. The results are shown in Figures 2 and 3.
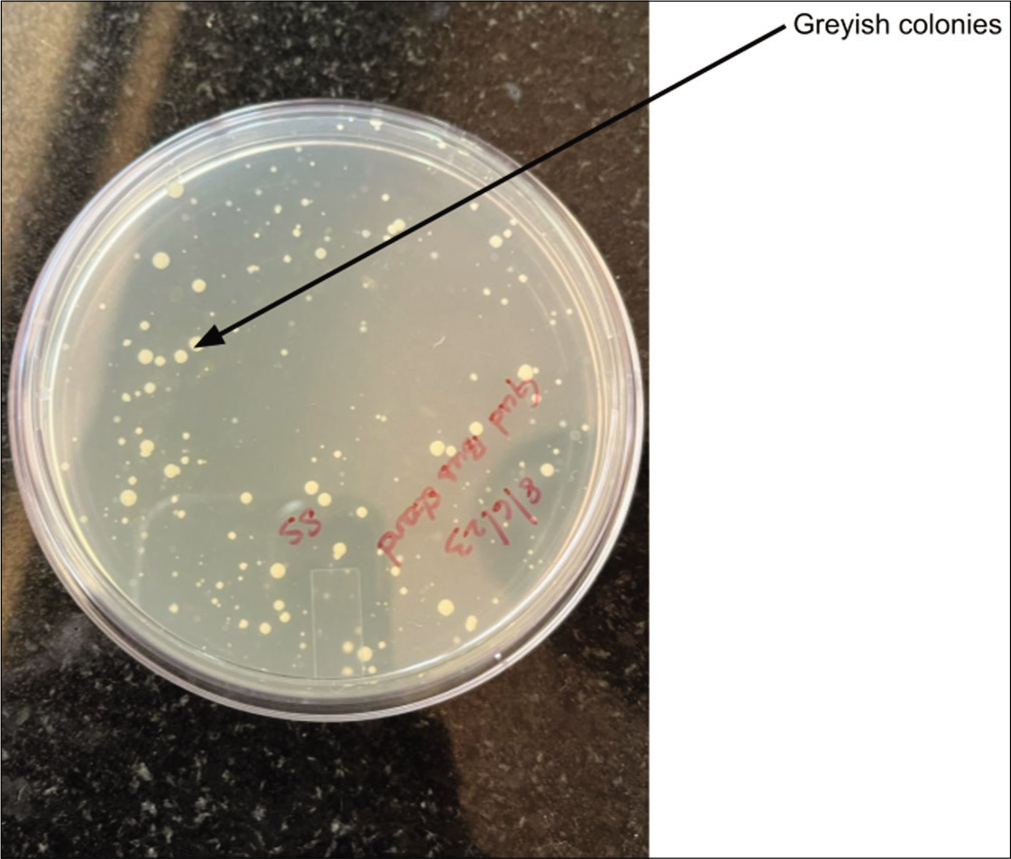
- Ethyl violet azide plate with greyish white colonies.
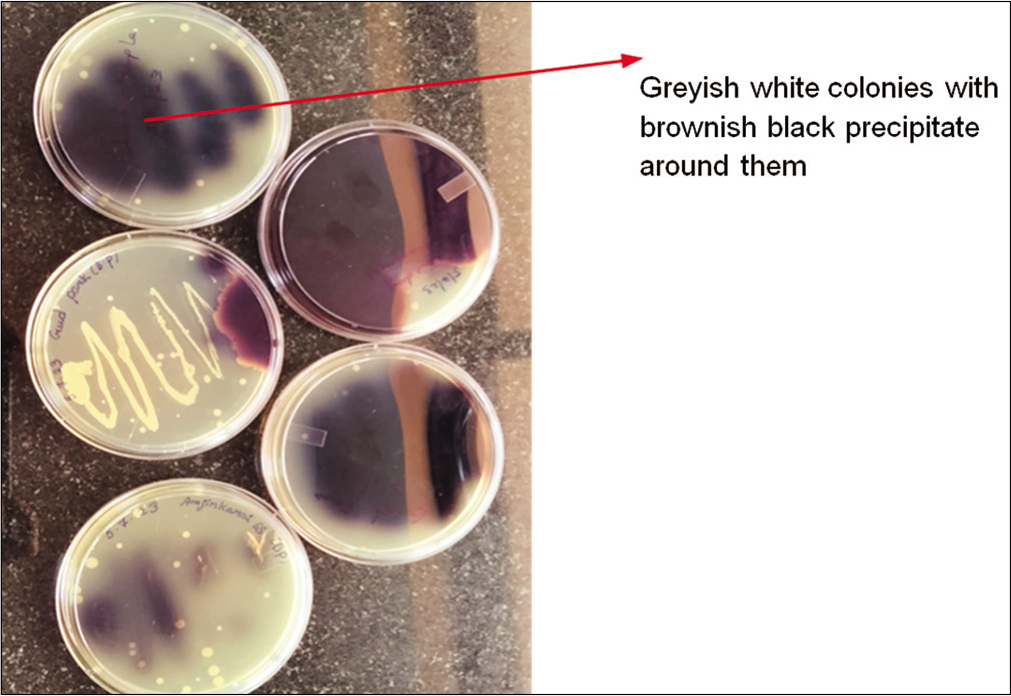
- Bile esculin plate with brownish-black precipitate colonies.
Bile esculin test
Colonies sourced from the EVA plates were streaked onto bile esculin agar plates, and the outcomes revealed a color change from dark brown to black. This transformation indicated the hydrolysis of bile salts within the medium.
Molecular detection of E. faecalis
The isolates underwent PCR analysis using SSOPs for the 16S rRNA gene. The anticipated amplicon was consistently observed across all isolates. Documentation of results was carried out using the gel documentation system. Both the positive and negative controls demonstrated satisfactory outcomes, with ATCC 29212 serving as the positive control. A 2 Kbp DNA ladder was employed as a marker, with the lowest band at 500 bp. The expected amplicon size of 557 bp was evident on the 1.2% agarose gel for 11 isolates (41%), refer to Figure 4 for a visual representation of the results.

- 16S Ribosomal ribonucleic acid polymerase chain reaction.
Detection of hyl gene in E. faecalis isolates
PCR targeting the hyl gene was conducted for all 11 isolates using SSOPs. Notably, the results indicated that none of the 11 isolates tested positive for the presence of the hyl gene. The results are shown in Figure 5.

- Hyaluronidase gene polymerase chain reaction.
DISCUSSION
E. faecalis, a versatile pathogen implicated in various human diseases, including urinary tract infections, bacteremia, infective endocarditis, meningitis, intra-abdominal infections, and wound infections, poses a significant public health concern. This study aimed to investigate the prevalence of E. faecalis in environmental samples collected in and around Chennai, shedding light on its distribution and the occurrence of one of its key virulence factors, the hyl gene. The comprehensive environmental sampling strategy, involving 32 diverse locations, revealed a substantial 41% incidence of E. faecalis. This prevalence underscores the widespread presence of the organism in the sampled areas. However, the intriguing finding was the absence of the hyl gene in any of the isolates from these environmental samples. This emphasizes a clear distinction between the environmental isolates and those associated with various diseases, particularly dental conditions. Contrastingly, in disease-related isolates, especially from dental caries, a high incidence of the hyl gene (72.4%) was observed. This stark difference in hyl gene prevalence between environmental and disease-related isolates points toward the pathogenicity and virulence factor specificity of E. faecalis in different contexts. Our findings align with previous studies that have reported the absence of the hyl gene in environmental samples. Studies conducted in regions such as Southwestern Iran, California, Puerto Rico, and poultry environments have reported varying occurrences of the hyl gene in E. faecalis isolates.[9-12] The current study, in harmony with these reports, strengthens the notion that the hyl gene is more commonly associated with clinical isolates, particularly those linked to oral health conditions. Moreover, the investigation of biofilm formation and vancomycin-resistant genes in Enterococcus faecium from clinical and environmental specimens has revealed a connection between virulence genes and biofilm formation. However, our study distinctively focuses on the absence of the hyl gene in environmental samples, contributing to the understanding of its distribution and significance in different settings. While our study provides valuable insights into the prevalence of E. faecalis and the absence of the hyl gene in environmental samples, the limitation lies in the relatively small number of isolates investigated. Future studies could expand on this research by including a larger sample size and exploring other potential virulence factors associated with E. faecalis in environmental settings.
In summary, our study contributes to the growing body of knowledge on E. faecalis, emphasizing the distinctive prevalence patterns of the hyl gene in environmental versus disease-related isolates. This understanding not only aids in the development of effective screening methods for environmental isolates but also underscores the potential health implications associated with the presence or absence of the hyl gene in different contexts.
CONCLUSION
Although our study identified a substantial prevalence of E. faecalis in environmental samples in and around Chennai, the incidence of hyl gene is low, necessitating further investigation with a higher number of samples. The absence of the hyl gene in these isolates challenges its conventional ubiquity, suggesting a potential divergence in pathogenicity between clinical and environmental strains. These findings emphasize the need for ongoing research to comprehend the specific dynamics of E. faecalis in diverse settings, informing targeted public health strategies.
Acknowledgement
Authors wish to thank the University management for providing the facilities required to carry out the research.
Authors’ contributions
DR, AS: Sample collection, analysis, manuscript preparation; SV: Ethical clearance, manuscript preparation; B PC: Work plan, design, manuscript preparation.
Ethical approval
The research/study was approved by the Institutional Review Board at Sri Ramachandra Institute of Higher Education and Research, number CSP/23/JUN/130/540, dated 30th January 2023.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript, and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Enterococcus faecalis in secondary apical periodontitis: Mechanisms of bacterial survival and disease persistence. Microb Pathog. 2023;183:106337.
- [CrossRef] [PubMed] [Google Scholar]
- The Enterococcus A model of adaptability to its environment. Clin Microbiol Rev. 2019;32:e00058-18.
- [CrossRef] [PubMed] [Google Scholar]
- Complete genome sequence of Enterococcus faecalis LD33, a bacteriocin-producing strain. J Biotechnol. 2016;227:79-80.
- [CrossRef] [PubMed] [Google Scholar]
- Hyaluronidases of gram-positive bacteria. FEMS Microbiol Lett. 2000;183:201-7.
- [CrossRef] [PubMed] [Google Scholar]
- Heat treatment of bacteria: A simple method of DNA extraction for molecular techniques. Kuwait Med J. 2009;41:117-22.
- [Google Scholar]
- The use of multiplex PCR reactions to characterize populations of lactic acid bacteria associated with meat spoilage. Lett Appl Microbiol. 2000;31:129-33.
- [CrossRef] [PubMed] [Google Scholar]
- Specific enzymatic amplification of DNA in vitro The polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263-73.
- [CrossRef] [PubMed] [Google Scholar]
- Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289-91.
- [CrossRef] [PubMed] [Google Scholar]
- Virulence genes among Enterococcus faecalis and Enterococcus faecium isolated from coastal beaches and human and nonhuman sources in Southern California and Puerto Rico. J Pathog. 2016;2016:3437214.
- [CrossRef] [PubMed] [Google Scholar]
- Biofilm formation capacity and presence of virulence factors among commensal Enterococcus spp. from wild birds. Sci Rep. 2019;9:11204.
- [CrossRef] [PubMed] [Google Scholar]
- Occurrence of virulent multidrug-resistant and in the pigs, farmers and farm environments in Malaysia. PeerJ. 2018;6:e5353.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of biofilm formation and vancomycin-resistant genes among isolated from clinical and environmental specimens in Lorestan hospitals. Iran J Microbiol. 2018;10:74-81.
- [Google Scholar]






