Translate this page into:
Evolution of the Indian population through the ages
-
Received: ,
Accepted: ,
How to cite this article: Iyer AR, Parvathi VD. Evolution of the Indian population through the ages. Sri Ramachandra J Health Sci 2022;2:47-55.
Abstract
The Indian subcontinent is synonymous with increased genetic diversity essentially attributed to migratory forces. Many theories that provide different perspectives regarding the current nature of the Indian subpopulation have been proposed. Each of these theories is equally insightful albeit contradictory in nature. Thus, there is ambiguity in terms of tracing the course taken by individuals forming the Indian subpopulation. A comparative analysis of the pre-existing hierarchal caste system and primitive Europeans, matrilineal factors, and increased incidence of population bottlenecks gives an insight into plausible theories that could have led to infringement across the borders of the Indian subcontinent. The Aryan exodus forms the crux of various migratory theories along with analysis of ancient DNA and Y-chromosome which are indicative of external infusion into the Indian gene pool. Thus, this review is a retrospective analysis based on concrete evidence generated from studies undertaken to map the evolution of population in the Indian context.
Keywords
Population genetics
Indian subcontinent
Aryans
Gene pool
Migration
INTRODUCTION
The Indian subcontinent is rich in terms of genetic diversity which can be attributed to migratory events that took place over the course of history. Due to the high degree of genetic variation, it has often been extremely difficult to trace the past of the individuals native to the Indian subcontinent. There are a variety of theories that have been expounded on to throw some light on the ancestry of various population groups that exist in India today. The most common theory that exists is the Aryan Invasion Theory (AIT) proposed by Max Muller in the 19th century, which suggests that dark-skinned Dravidians (people belonging to the Indus Valley Civilization [IVC]) were native to the Indian subcontinent before the Indo-Aryan invasion, westerners of European origin, and fair skin, who spoke Vedic Sanskrit. The Indo-Aryans subsequently subjugated the native Dravidians and drove them south post which they established the rudiments of present-day Hinduism and the hierarchical caste system. This theory was, then, re-named to be called the Indo-Aryan Migration Theory which stated that the Indo-Aryans migrated to the Indian subcontinent instead of invading it, but the consequences of their actions remained the same. Although, a theory opposing these facts also gained a lot of popularity among the masses and it was called the Indigenous Aryans Theory or the Out-of-India theory. It stated that the Indo-Aryans and their languages originated in the Indian subcontinent and the IVC was a Vedic Civilization and not a Dravidian Civilization as proposed by the AIT. The debate regarding the origin of the Indian subcontinent has been going on for over a decade, ending with ambiguous statements and assumptions. The only plausible method by which one can obtain definitive answers is by resorting to scientific approach:
Analyzing the DNA obtained from archaeological sites with the help of scientific techniques, testing for certain markers that have been evolutionarily conserved over time.
Resorting to systematic observations and conducting replicable tests based on hard evidence to come to a conclusion.
The aim of this review is to pool all the information regarding the history of the Indian populace and accordingly infer as to how the native Indians came into being and what shared ancestry they represent.
A LINK BETWEEN THE HIERARCHICAL CASTE SYSTEM AND PRIMITIVE EUROPEANS?
The Indian subcontinent is extremely populous and there is tremendous diversity that exists in terms of cultural, linguistic, and genetic aspects. The only continent to outdo India in this regard is Africa. A variety of groups native to the Indian subcontinent phenotypically resemble Africans, Europeans, Southeast Asians, etc. Moreover, a vast majority of individuals arise from the amalgamation of two divergent ancestral lineages, namely, Ancestral North Indians (ANI), related to West Eurasians, and Ancestral South Indians (ASIs), distantly related to the islanders native to Andaman. The diversity that exists can be credited to migratory processes that began as early as during the Late Pleistocene to prehistoric period.[1] There exists tangible evidence regarding rearrangement of population within the subcontinent itself due to activities such as changes in the economy and the advent and spread of agriculture.[1]
The four main castes include (from the upper caste to the lower caste), Brahmins (priests or the clergy), Kshatriyas (warrior class), Vaishyas (business class), and Shudras (menial worker class). In addition to Hinduism, other religions practiced by the populace include Christianity, Islam, Buddhism, Sikkhism, and Judaism.[2] A study conducted by Bamshad et al., in 2001, states that the affinity a particular caste has to Europeans is proportional to the order, in which it appears in the system of hierarchy, wherein the upper castes are more similar to East Europeans in particular. The lower classes on the other hand exhibit lesser similarity toward the European ancestry and more similarity toward Asians.[3] This study backs the fact that the Indo-Europeans migrated into the Indian subcontinent, entrenched the caste system, and subsequently inducted the people native to the subcontinent into the caste fold. Although the need to investigate further is imperative as the samples were derived from a particular geographical area and extrapolating the data obtained from a particular data set to match an entire population belonging to the Indian subcontinent needs extensive validation[3,4] [Figure 1].
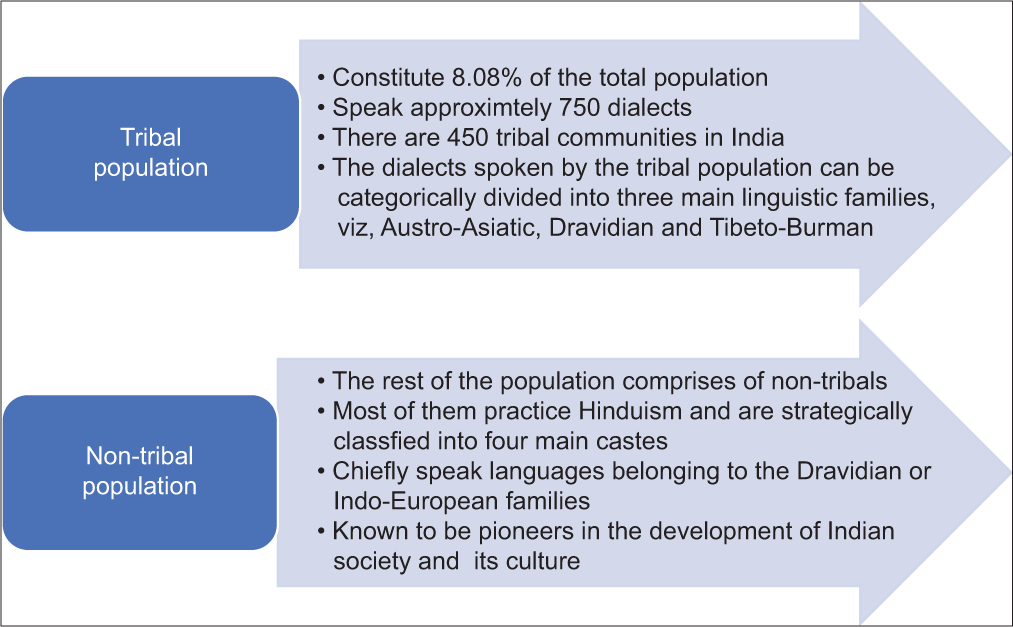
- The differences between the existing tribal and non-tribal population. The differences between tribal and non-tribal population are established based on the percentage of population constituted by each, dialects or languages spoken and their subsequent bifurcations into other groups or communities (Singh K.S., 1992, Kosambi, 1991).
MATERNAL LINEAGE: A DECIDING FACTOR
Data obtained post studying 58 DNA markers, including but not limited to – autosomal, Y-chromosomal, and mitochondrial (mt) in ethnically diverse population belonging to the Indian subcontinent, were statistically analyzed to provide genomic evidence regarding a multitude of research questions. The data analyzed were construed in the following way: The intrinsic unity of female ancestries in the Indian subcontinent is a consequence of the probability that the number of female settlers in the initial period was small in proportion; differentiation among the caste and tribal population was undoubtedly high; The Austro-Asiatic tribals were primary settlers on the subcontinent, thus supplementing one anthropological theory and debunking others; the northeast was the gateway through which a majority of humans entered the Indian subcontinent; substantial genetic congruity shared by Tibetan-Burman tribals; and Austro-Asiatic tribals supports the hypotheses of the existence of a communal habitat shared by both in Southern China. Although, differentiation between the two groups can be established on the premise of Y-chromosome haplotypes; the Dravidian tribals, although predominantly spread out across the Indian subcontinent before the appearance of the Indo-European-speaking nomads, quickly withdrew to the southern pockets of India to escape dominance; the advent of newer population as a result of fission has resulted in drift and founder effects, which have left their marks on the genetic fabric of the present-day population; the genetic affinity of upper castes with the population found in Central Asia is substantially similar, although upper castes native to Northern India are more similar to population found in Central Asia as opposed to upper castes found in Southern India; and genetic histories of the present-day population have been entirely exterminated as a consequence of the historical gene flow into the Indian subcontinent. As a result, of which there exists no clear consonance with sociocultural, geographical, or genetic affinities.[4]
The genetic variation in 44 ethnic population that exhibit contrast in terms of geographical, social, and linguistic interests was analyzed in a study conducted by Bamshad et al., in 2001. Ten restriction site polymorphisms (RSP), one insertion/deletion, and 1 hypervariable segment on mitochondrial DNA (mtDNA) were studied. The data obtained post-analysis of the Hpa1 np 3592 mtDNA restriction site locus were suggestive of the existence of 32 definitive polymorphisms present on the ten-locus mtDNA RSP haplotypes (maximum number of restriction polymorphisms was found in Rajput and Tipperah, close to 13, whereas a minimum of two were found in Kota and Toda) among 1490 individuals that were studied from 44 population, out of which 5 (15.7%) were exclusive to population found in the northeast. The frequency with which the haplotypes were distributed in a population did vary greatly, although one among the multitudes that were analyzed was found to be a part of almost 46.4% of all the mtDNA molecules. This haplotype was termed as haplogroup (HG) M. Population exhibiting reduced occurrence of HG-M belonged to the ethnicity comprising northeastern groups (Mog, Jamatiya, Chakma, and Toto) and northern groups (Punjab Brahmins, Muslims, Rajputs, and Uttar Pradesh Brahmins). The haplogroup predominantly prevalent in the aforementioned groups was HG-U. Regardless of HG-M not being primarily prevalent in abundance in the latter groups, it was the second most abundant haplotype over-all in the northern and northeastern groups[4] [Figure 2].
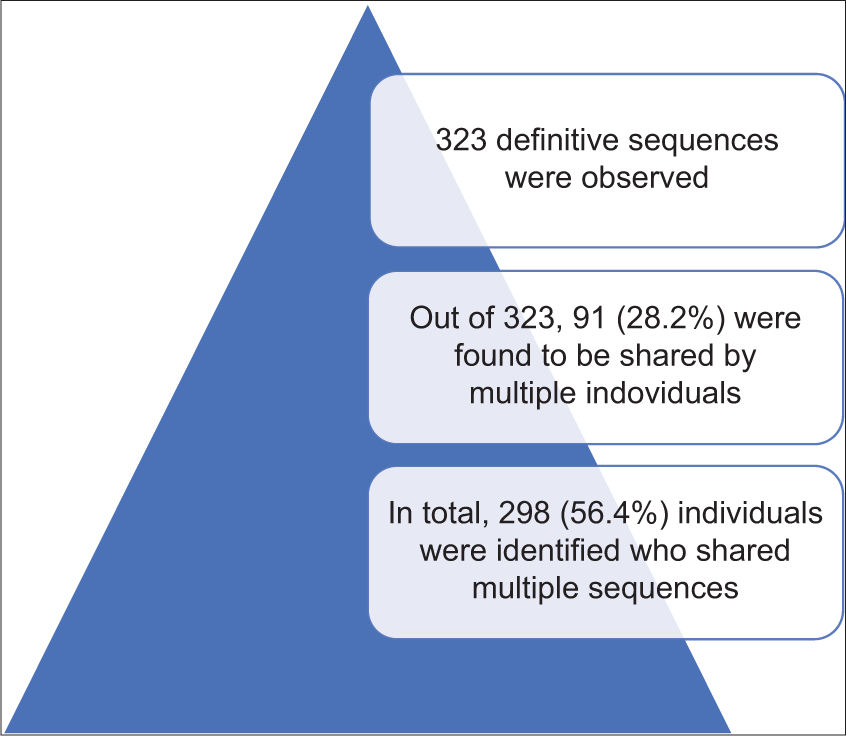
- Sequential representation of data obtained post-analysis of hypervariable segments of mtDNA. Comprises the results obtained post-analysis of hypervariable segments on mtDNA taken from 528 individuals. It indicates the number of similar sequences found on all 528 individuals and the number of distinctive sequences. The number of individuals sharing shared sequences was also tabulated (Basu et al. 2003).
In conclusion, the distribution of mtDNA haplotypes in the population is indicative of the presence of strong female lineages in the Indian subcontinent. This could be attributed to the fact that only a small proportion of these founding female lineages existed in India due to founder effect caused due to few women entering the Indian subcontinent during the process of migration or the fact that the founding females derived from an ancestral population irrespective of the size or number possessed uniform mitochondrial haplotypes.[4] Hypervariable segments on mtDNA for 528 individuals were analyzed. The data obtained was as follows: [Figure 2].
THE EARLY INHABITANTS OF INDIA: A PLAUSIBLE THEORY
Linguistic and sociocultural evidence assimilated over the years is suggestive of the fact that Austro-Asiatic tribals might be the indigenous inhabitants of the Indian subcontinent.[5-7] Alternatively, there are reports formulated by other scholars that suggest that tribal groups conversing in Austro-Asiatic and Dravidian languages are evolutionary by products of protoAustraloids which were considered to be the predecessor.[8] Furthermore, these conflicting reports also go on to say that Tibeto-Burman speaking tribal groups immigrated from Myanmar and Tibet at a later date.[9] The reason Austro-Asiatic tribal groups are considered as the oldest and most primitive inhabitants of the Indian subcontinent are because they possess an abundance of ancient East Asian HG-M mtDNA. Furthermore, they also exhibit immense diversity of hypervariable segment 1 (HVS1) among all the other subHGs due to which they are touted as the earliest settlers. Although all the population belonging to different socialistic groups have shown significant population expansion in terms of distribution HVS1 mismatch, the relevant statistics state that the Austro-Asiatic group of tribals has shown tremendous when compared to other groups, almost equivalent to 55,000 years. It is 15,000 years greater than the existing estimates for other groups.[10] Nevertheless, there is ambiguity that exists regarding, whether or not, the expansion took place in the Indian subcontinent or elsewhere. The existence of a relatively juvenile subgroup of a haplogroup called the M4 lacks documentation among the Austro-Asiatic tribal group. The merging time of this subgroup is estimated to be 32,000 ± 7500 years before present and the total frequency is estimated to be approximately 15% in the Indian subcontinent. In conclusion, M4 might have arisen post the entry and expansion of the Austro-Asiatic tribal group in India.[4]
THE ARYAN EXODUS
Indian history has witnessed a series of events that have led one to believe an array of facts revolving around IndoEuropeans who called themselves the Aryans. The most pertinent question that has remained unanswered over so many decades is that whether or not the Aryans streamed into the Indian subcontinent around 2,000–1,500 BC post the downfall of the IVC, ushering in a set of idiosyncratic cultural practices and the rudiments of Sanskrit. Contrary to popular belief that, there was minimal external inculcation in the Indian gene pool over the past 12,500 years or so (due to availability of only matrilineal DNA data, where genetic information is transmitted from parent to offspring), there have been recent findings related to Y-DNA that seems to suggest otherwise. Solid evidence regarding external introduction of genes into the Indian male lineage during the same time period has been procured. Evidence suggests that a group of males harboring an Eastern European genetic structure invaded India many millennia ago through the Northwestern region of the subcontinent. The existence of genetic variations among the lower and upper castes provides evidence explaining how the caste system originated. Researchers state that the invaders and migrants who entered the subcontinent could have been the harbingers of the caste system. This could be attributed to the fact that these invaders and migrants were found to have descendants (who were predominantly males) largely belonging to the higher castes as opposed to the lower castes.
Lack of information regarding the emergence of the Central and South Asian population can be attributed to the absence of ancient DNA. To gain clarity regarding the same, an extensive study was carried out, wherein whole-genome ancient DNA data were generated from 523 individuals from northernmost South Asia and Central Asia starting from the Mesolithic period. The data thus obtained were then coanalyzed with previously published data regarding ancient DNA from all over Eurasia and data obtained from diverse contemporary population. The whole-genome data from ancient DNA were derived from 523 ancient individuals that had not been incorporated into the sample size previously. To increase the quality of the data generated, 19 individuals that were previously sequenced were also included. The individuals were procured in the following manner; (1) 132 from northern Pakistan, (2) 209 from the northern forest zone and the Steppe region (these constitute present day Russia and Kazakhstan), and (3) 182 from Turan – Southern part of Central Asia (present-day Kyrgyzstan, Afghanistan, Tajikistan Turkmenistan, and Uzbekistan) and Iran. These ancient individuals belonged to the Iron, Copper, Bronze and Mesolithic Age Turan and Iran, some of them were early hunter gatherers using ceramic tools from the forest zone in Western Siberia, others were Copper and Bronze Age pastoralists from the central Steppe region; individuals belonging to the Bronze Age Kazakhstan; and historical settlements found in Chitral and Swat districts of modern-day Pakistan were also a part of the sample size.[11]
Post sequencing, the West Siberian Forest zone belonging to the Steppe category highlighted three individuals who had major roles to play in this study coming to fruition. They were found to be mixtures of combinations of ancestry unknown to have had contact, namely, 30% West Siberian, 50% from Ancestral North Eurasians, and 20% related to the East Asians that we know today.[12,13] The Indian scenario shows dissimilar fractions of West Eurasian associated ancestry in South Asia having originated from reassembled population of ANI and ASI, wherein the mixture was said to have occurred post 2000 BCE.[14,15] Over time, analysis revealed that the roots of the Indian Cline go deeper than one had previously proposed, whereby they could be traced back to three primary ancestral groups, namely, the early Iranian agriculturalists, Steppe_EMBA (from European Middle Neolithic Agriculturalists), and Onge. Moreover, since ancient DNA belonging to the individuals buried in accordance with the cultural contexts of the society is unavailable, one cannot dismiss the possibility that the group represented by these outlanders (belonging to the isolated Swat valley of the IVC known to share similar ancestral profile as those belonging to Ancient Ancestral South Indians (AASIs), the early Iranian agriculturalists and the West Siberian Hunter Gatherer) called the Indus Periphery was confined to the northern borders and was not archetypal of the ancestry of the complete IVC. Although heterogeneity did exist among the people belonging to the IVC, wherein one individual belonging to the Indus Periphery exhibited close to 42% AASI ancestry, whereas the other two had a lower percentage of AASI ancestry, close to 14–18%. Nevertheless, they were mixtures of the AASI and Iranian agriculturalist-related ancestry. This essayed a significant role in the formation of South Asia.[11]
Evidence suggests that the Iranian-agriculturalist-related underwent a genetic mixture with AASI somewhere between 4700 and 3000 BCE. Hence, it must have integrated itself into the IVC by the 4th millennium BCE. There are possibilities that the Iranian agriculturalist-related ancestry might have been predominant in South Asia even before the stipulated time period. This can be attributed to activities like cultivation of wheat, barley and herding of sheep and goats that made an entry in the 7th millennium BCE.[16,17] Corroborative evidence regarding the same has been identified in the hills surrounding the IVC at sites like Mehrgarh. Irrespective of the time period announcing the arrival of these agricultural species, genetic data validates that the individuals associated with the Indus Periphery ancestry formed a large part of the ANI and ASI. These groups were instituted around the 2nd millennium BCE, around the same time, the IVC was in decline along with major settlement changes in the northern portion of the Indian subcontinent. A lesser-known hypothesis states that movement of the Steppe_MLBA groups down south and their subsequent mixture with the Indus Periphery-related groups led to the formation of the ANI; at the time, the IVC was in decline. Alternatively, movement of other Indus Periphery-related groups further down south and east to engage in genetic mixture with the AASI groups inhabiting the peninsular region of the Indian subcontinent led to the emergence of ASI. Therefore, this theory validates suggestions regarding the dispersal of Dravidian languages being a consequence of the spread of the IVC. Although scenarios propagating the fact that Dravidian languages are derivatives of the pre-Indus languages of the peninsular region exist, there could be some truth in them due to the fact that the AASI ancestry is the progenitor of the ASI ancestry.[11,17]
Regarding the origin of Indo-European languages, it has been found that a vast majority of Indo-European speakers residing in Europe and South Asia show significant similarity in ancestry to the Yamnaya Steppe pastoralists (Genetically similar to the Steppe_EMBA cluster), thereby insinuating that “Late-Proto-Indo-European” which is considered as the progenitor of all modern Indi-European languages was akin to what the Yamnaya individuals used to communicate.[11]
INDO-IRANIAN HOMELAND
Kodumanal is a site located near one of the tributaries of the Kaveri River called the Noyyal tributary. It is touted as an exceptional site as the Late Iron age and Early Prehistoric rudiments were found to be based here. Big stone sculptures built to honor the dead were characteristic to this era. Contrastingly, accommodation for the villagers usually comprised temporary huts made of mud and leaves. They also had an inclination toward fine pottery, iron-production, and farming. Artifacts belonging to the Megalithic era, when unearthed from peninsular India (stretching from Vidarbha located in Central India to the Thamarabharani river located deep down South) showed the presence of similar weapons and tools. This could be attributed to the fact that iron smelting was an occupation that was rampant in Central India. The term Agaria was given to individuals belonging to this field of work. It was discovered later that the name Agaria referred to “a group of iron smelters.” The individuals in this group did not share common ancestry, names, or customs, although their technology and mythology were found to be similar. Mitannians were known to be native to Iran. The most intriguing part of their existence was the striking similarity their local language had to Vedic Sanskrit. Burrow (1973) proposed a common homeland that existed in the Hindukush uplands. He ascertained that it was highly possible that a few groups broke away and migrated toward India and Syria. The migration was said to be gradual, post which it was succeeded by horse-mounted invasions. Furthermore, there have been instances wherein horse skeletons, horse strappings, or lances spanning meters that could only be used by a horse rider have been found in large numbers across the entirety of the Vidarbha region (150 km from west to east) that exists in the Indian subcontinent.[18]
GENETIC MIXTURE AND POPULATION BOTTLENECKS
In 2009, a study conducted by Reich et al. involving the analysis of 25 different groups of the Indian population, distinctly described how there was an evident genetic mixture of two ancestral groups, namely, ANI and ASI. The former is related to Central Asians, Middle Easterners, Caucasians, and Europeans, whereas the latter primarily belong to the subcontinent. To gain more clarity regarding the same, the sample size was increased from 25 to 73 Indian groups. At the beginning when there was genetic mixture of people belonging to the ANI and ASI descent, the segments were estimated to span the entire length of the chromosome. Possibilities of fragmentation of these segments at one or two places per chromosome per generation have led to shortening of these segments during recombination of paternal and maternal genetic material during production and post-fusion of the egg and sperm. Post measuring the length of the ANI and ASI segments in the genomes of the sample group, it was ascertained that the age of the population mixture varied between 1,900 and 4,200 years based on the type of population subjected to this analysis.[19]
Over the years, there have been plausible theories backed with evidence, which state that there have not been groups in the Indian subcontinent that has been free from genetic mixture, although research suggests that the groups down south have older dates as compared to the groups that inhabit the north. This could be attributed to the fact that the northern groups have multiple mixtures. Genetic data establish the fact that no mixture in the population existed close to 4000 years ago. Post this time period, a ubiquitous mixture plagued every group in the subcontinent. Discreet tribal groups which thrived in isolation were also subjected to population mixture. The fact the Indian population has evolved from a random mix of various groups is suggestive that social hierarchy like the caste system did not exist. It was founded and established recently due to which endogamy was widely propagated, thus freezing everything in place. The racial theory states that the advent of the caste system was observed after the arrival of the Aryans in the Indian subcontinent. The premise of the same was based on differentiating the Aryans from the non-Aryans on the basis of their physical features, religious practices, speech, and complexion. The ensuing endogamy, thus, ensured the preservation of the aforementioned demarcation.[17,19]
A large fraction of Indian groups that exist today are the result of population bottlenecks. This occurs when a small number of individuals have many offspring and, in turn, their descendants have a larger number of progeny, those that are isolated from the surrounding population either due to geographical or social barriers. Famous examples include, people of European ancestry contributed to most of the ancestry of the Finnish population (around 2000 years ago) and the Ashkenazi Jews (around 6000 years ago). These population bottlenecks are, often, subjected to afflictions characteristic of recessive disorders. Recessive disorders can manifest only when identical copies of recessive allele carrying the mutation are present on either chromosome. The percentage of individuals exhibiting this condition is usually 25% in a population. Due to population bottlenecks, the frequency of an individual carrying a recessive allele and passing it on to the future generation increases manifold due to the possibility of the offspring inheriting recessive copies from both the parents.[20]
Due to the numerical vastness of the Indian population, bottlenecks were a common phenomenon. An estimated one-third of the Indian jati groups has been found to descend population bottlenecks that are either equally strong or stronger than what occurred in the Finns or Ashkenazi Jews. The caste system in India is referred to as “jati.” There exist close to 3000 jatis in India, all of which cannot be classified with regard to status using a singular, uniform ranking system. Jati groups are widespread throughout the Indian subcontinent and are a direct consequence of the arrival of the Aryans. The most startling discovery in terms of population bottleneck was an ideology called the Vyasa, peculiar to the state of Andhra Pradesh. It comprised approximately five million individuals whose population bottleneck could be dated back to 2000–3000 years (ascertained through analysis and measurement of segments common to individuals of the same population). The existence of such a strong bottleneck through centuries was indicative of strict endogamy, wherein no genetic mixture was allowed as even an average influx of 1% of genetic mixture per generation could have eradicated the rudiments of the population bottleneck altogether. Another interesting fact about the Vyasa is that they did not thrive in isolation, in fact, they lived in close proximity to other clans and groups. Their strict rules regarding endogamy along with religiously passing on cultural practices and beliefs from one generation to the next transformed them into one of the most famed bottlenecks in history. An example of a recessive condition being rampant in individuals belonging to the Vyasa ancestry is increased rate of muscle paralysis in response to muscle relaxants administered before surgery. As a result, clinicians refrained from prescribing drugs inducing such effects in people belonging to the Vyasa lineage. This condition is caused by low levels of the protein butylcholinesterase in certain Vyasa individuals. Post-analysis, the genetic data procured ascertains that the frequency of this recessive mutation is close to 20% in Vyasa individuals which are estimated to be far higher than what is observed in all the other Indian groups. This could be attributed to one of the Vyasa founders carrying this mutation. About 4% of the Vyasa population carry two copies of the recessive gene, thereby harboring increased susceptibility to disastrous consequences.[20]
ANCIENT DNA (ADNA): AN ELUCIDATION?
Unfortunately, the absence of investigable ancient biological remains in the Indian subcontinent makes it an arduous task for researchers and scientists to trace and understand the past. Nevertheless, archeological evidences that indicate the existence of hominins in Andhra Pradesh, in the Jurreru river valley, do exist. Although, prior to and post the upsurge of the Youngest Toba Tuff, there is comprehensive absenteeism of biological remains. The oldest substantiation of a prehistoric settlement indicating the rudiments of modern-day human civilization is the Mesolithic settlement, established in the Gangetic plains. This essentially dates back to 10,000 years before what one can see today. Post the Mesolithic civilization, the Harappan civilization gained impetus and spread all over the northwestern portions of the Indian subcontinent. The process of analyzing these Harappan remains are underway as they would play a pivotal role in stringing together information regarding the migratory, settlement, and disease patterns that existed in the past. Thus, analysis of aDNA is considered a discipline that is gaining importance over the last decade. aDNA could be inspected in various forms, including but not limited to, museum specimens, archeological materials, and other biological substances.[1]
The propinquity between the people belonging to the IVC and the Gangetic plains and also the modern-day Indians along with their association with pre-existing cultural practices that existed all over Asia before their subsistence has been an open-ended question throughout the course of Indian history. There is great potential in analyzing the ancient Harappan DNA and comparing it with that unearthed from the Megalithic burials which are concerted more toward the Southern part of the Indian subcontinent. Due to technological advancement and significant progress in modern DNA database analysis, researchers can ascertain and validate an outline of the South Asian population demographic that existed prehistorically. The data obtained post-analysis of aDNA can be fruitfully used to reconstitute the structure of South Asians that did exist, accurately, as they are chronicled as “pockets of endogamy.” Furthermore, the data generated from aDNA post-inspection would also aid in establishing and accentuating the genetic correlation between the Harappan culture and the Bronze age cultures characteristic to South India that arrived at a later time period. It could also be suggestive of the plausible relationship between the Harappans and modern-day Indians.[1]
FOUNDER EFFECT
It is characterized by reduced genetic diversity which occurs when a population is descended from a smaller group of colonizing ancestors. The Indian subcontinent being highly populous exhibits an array of population bottlenecks which increase its susceptibility to reduced genetic diversity primarily facilitated by strict endogamy. For example, Hydatidiform moles (HM) is a condition, wherein the pregnancy is rendered abnormal due to certain abnormalities in embryonic development or the absence of the same in entirety. Hydropic degeneration is another phenomenon that accompanies this condition. The rate of incidence of this condition is ideally one in every 1000–1500 pregnancies among Westerners. The frequency of the same in the Indian population is quite high, one in every 83–500 pregnancies. Ideally, HMs are not recurrent, they are usually sporadic though possibilities of women with sporadic moles developing a second mole at a later stage does exist. Statistics state that an average of 1–6% of women is susceptible to developing second moles, whereas another 10–20% of women might suffer spontaneous abortion as a consequence, also termed as non-molar reproductive wastage. Genomic analysis of women with recurrent Hydatidiform moles has identified mutations in the gene NLRP7. As of now, there are 23 mutations that have been identified in the NLRP7 gene, of which four of them have been identified in families that are unrelated and from the same population, thus indicating the presence of Founder effects for these mutations. Being a recessive disorder, two copies of the mutated gene are required for the condition to manifest. Studies have shown that these mutations have no effect whatsoever on male reproduction. The presence of these mutations in a large number of Indian groups is indicative that there is reduced genetic diversity, although it is advantageous as this condition prevents the reproduction of homozygous females by inducing abnormal developing and thus spontaneous abortion thereby favoring genotypes that are heterozygous in nature. It is an interesting aspect as the future generation is at lesser risk of experiencing the brunt of this condition.[21]
An example of founder effect mutations that have disastrous consequences in Indian groups includes a founder variant that dramatically increases the incidence of FANCL Fanconi anemia in the India population. Fanconi anemia is caused due to mutations in one of the 22 genes involved in pathways associated with DNA repair that aid in removing inter-strand crosslinks. An E3 ubiquitin ligase, FANCL, plays a pivotal role in the DNA repair pathway. Thus, mutations in the gene coding for this enzyme give rise to a founder variant that harbors the potential of increasing the risk of incidence of FANCL Fanconi anemia in the Indian population. Ideally, these variants are extremely rare, wherein only 7–9 cases are reported annually worldwide. Genetic analysis has proven that this founder variant is rampant in individuals sharing a South Asian ancestry and that Indians show an increased likelihood of developing this condition.[22]
SEX BIAS
In comparison to autosomes, the X-chromosome is a part of a smaller population, wherein its effects are discernible. Due to this, it is subjected to stronger waves of random genetic drift and recombination. Moreover, the mutation rates associated with the X-chromosome are considerably low. In addition to this, the X-chromosome exhibits a higher selection pressure in males. Nonetheless, it provides constructive information and evidence regarding the existence of sex bias. To generate data to substantiate the claims made, 107 females were chosen at random from 15 mainland population belonging to the Indian subcontinent. Analysis revealed a sex bias was prevalent in all ancestries except Ancient Ancestral Asians. On the other hand, Ancient Tibeto-Burman exhibits an overload of constituents belonging to the X-chromosome in comparison to autosomes. Conversely, the ANI component exhibits the opposite.[23]
Y-DNA INSINUATING EVOLUTIONARY EVIDENCE OF EXTERNAL INFUSION IN THE INDIAN GENE POOL
At present, there are four major linguistic families that inhabit the Indian subcontinent, namely, Austro-Asiatic, Indo-European, Dravidian, and Tibeto-Burman. There are assumptions that their entry into the Indian subcontinent might have been at different time periods. Out of the aforementioned linguistic families, Austro-Asiatic is considered to be one of the oldest, having a variety of linguistic features, and considerable branching of their noun forms. This category has three sub-types, namely, (a) Mundari primarily spoken by the inhabitants of the Chota-Nagpur plateau spread across Eastern and Central India; (b) Khasi-khmuic: it was believed to be a part of Mon-Khmer earlier but at present is characteristic only to Northeast India, home to the Indian Khasi sub-tribes, and (c) Mon-Khmer, spoken by tribes (Nicobarese and Shompen) inhabiting the Andaman and Nicobar Islands. The Mon-Khmer and the Indian Khasi tribes (to a large extent) show certain similarities in terms of features characteristic to the Dravidian linguistic family. Furthermore, except Mundari, the languages spoken by the other two tribes (Khasi-khmuic and Mon-Khmer) are not restricted to the Indian subcontinent, but are spoken by population inhabiting South Asia as well. Assumptions regarding the Indian subcontinent serving as a gateway for migrations to South Asia have been considered. Two migratory routes have been suggested based on archaeological, linguistic, and classic genetic marker, namely, from Africa to India through Central Asia and from Africa to Northeast Asia and then to the Indian subcontinent. Basu et al. discovered a haplogroup K-M9 which was found in a relatively high frequency in the Mundari population, thereby inferring that migration of the Austro-Asiatic population occurred from Africa to India through Central Asia which is essentially flawed as this haplogroup is predominantly present all over Asia and is significantly integrated in individuals native to East Asia.[24]
Due to availability of limited genetic evidence from mtDNA, genetic data post-analysis of mtDNA 9bp deletion and insertion polymorphism by Thangaraj and Rai and Prasad et al. presented that only east-Asian specific mtDNA haplogroups existed in Nicobarese tribes. Similarly, Roychoudury et al. and Metspalu et al. presented data that indicated that only Indian-specific mtDNA haplogroup existed in Mundari tribes. Inspection of the Y-chromosome Single Nucleotide Polymorphism and Short Tandem Repeats was undertaken to unearth the origin and subsequent expansion of the Austro-Asiatic population in the Indian subcontinent. Evidence suggests that a haplogroup called O-M95 arose in the Indian Austro-Asiatics individuals explicitly characteristic to the Mundari tribes, whose ancestors, then, moved across the subcontinent and migrated to shift base to places like Southeast Asia carrying the haplogroup along. In due course, the migratory process also witnessed the Mon-Khmer population shifting base from Southeast Asia to the Andaman and Nicobar Islands, thereby establishing shared ancestry between native Indians and the residents of Andaman and Nicobar.[24,25]
Anthropologists surmise that a few Negrito-hunter gatherer tribes native to the Andaman and Nicobar Islands (Jarawa and Onge) may play a pivotal role in discerning the ancestry of the people belonging to eastern and southern Asia post movement of humans out of Africa. Evidence suggests that few of the tribes native to Andaman and Nicobar Islands share a common ancestry with the ASI related groups and not with the ANI related groups. The genomic variation was over 13% which, thereby, differentiated the Jarawa and Onge tribes belonging to the Andaman and Nicobar Islands from the rest of the mainland. This was indicative of a prolonged separation and negligibility in gene flow.[23]
To throw some more light on Andaman and Nicobar Islands playing an important role in evolution of the Indian population, another study was conducted, in 2007. 2169 samples from 21 different tribal groups from varying parts of the Indian subcontinent were collected and scanned for the Y-chromosome Alu polymorphism. The results post-analysis stated that there was an increasing similarity (8–65%) of the Y Alu polymorphism (YAP) insertion between the Northeast Indian tribes and the Jawara samples collected from the locals belonging to the Andaman and Nicobar Islands during a previous study. Haplotype D was found to be the bridge of similarity between the two population. The frequencies of YAP that were previously reported in the population belonging to the mainland of the Indian subcontinent were close to negligible. This could be attributed to a commonly known factor called genetic drift, which serves as a plausible reason for varying degrees of YAP insertion in the mainland population. Conversely, founder effect could be accredited with increasing incidence of Haplotype D in the inhabitants of the Andaman and Nicobar Islands. Combinatorial analysis of results of YAP insertion and various studies regarding the mtDNA lineage is suggestive of early migration from Africa to Andaman and Nicobar Islands. Furthermore, findings associated with northeast Indian tribes showing similarities in haplotype are a major step toward discerning the evolutionary history of the Indian subcontinent.[25]
The Tibeto-Burman subfamily belonging to a parent family called Sino-Tibetan has essentially been bifurcated into the following groups: Bodic, Baric, Karen, and Burmese-Lolo. Post conducting a study of the Y-chromosome haplogroups, namely, p*, BR*, R1a, J, K*, and L, it was inferred that the Baric group relocated toward the south and occupied the northeastern region of the Indian subcontinent once they crossed the Himalayas. This occurred post the proto-TibetoBurman people abandoned the Yellow River Basin which was their homeland initially. It was found that this group showed the absence of the YAP insertion element. Similarly, the Tibeto-Burman individuals in India do not show the presence of the YAP insertion element either. These findings validate the fact that the Austro-Asiatic individuals entered the Indian subcontinent through the northeastern corridor. Khasi, a major tribal group belonging to the northeastern region, also converse in a dialect inherent to the Austro-Asiatic subfamily. Apart from India, population subgroups in Southeast Asia also communicate using dialects ancestral to the Austro-Asiatic tribal groups. Thus, it is safe to assume that a part of the Austro-Asiatic tribal groups did indeed enter from the northeastern corridor and set foot into the Indian subcontinent.[4]
CONCLUSION
Data collected from various studies that aimed to trace the evolutionary pattern of the individuals belonging to the Indian subcontinent state that there has been an enormous amount of genetic mixture due to integration of various migratory subgroups. The IVC was the hotspot of genetic integration, wherein the peripheral regions, post their decline, became optimal settlement areas to the Steppe_ MLBA groups that migrated Southwards. Furthermore, it has been established that the IVC formed a major part of the ANI (Ancestral North Indian) and ASI subgroups. Parts of Iran, Russia, and Europe were found to play a major role in the shared ancestry that exists in the modern-day Indians, we see today. Due to the fact that only mitochondrial DNA was assessed which was transmitted through the female lineage, it was assumed that there was no external infusion into the Indian population, but recent studies on Y-chromosome assessment have refuted the aforementioned facts, thereby suggesting that the Aryan exodus was a reality. The Andaman and Nicobar Islands have also been found to play an important role regarding the establishment of shared ancestry with the northeastern states. Furthermore, migration has been found to occur primarily through the northeastern corridor of the Indian subcontinent. Post migration, settlement, and population expansion of the westerners into the Indian subcontinent, endogamy was subsequently propagated which led to population bottlenecks and founder effect for various kinds of genetic phenomena, thereby adding to the genetic diversity.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Linguistics and the study of early Indian history In: Thapar R, ed. Recent Perspectives of Early Indian History. Bombay, India: Popular Prakashan; 1995. p. :53-79.
- [Google Scholar]
- Genetic evidence on the origins of Indian caste population. Genome Res. 2001;11:994-1004.
- [CrossRef] [PubMed] [Google Scholar]
- Ethnic India: A genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277-90.
- [CrossRef] [PubMed] [Google Scholar]
- The language heritage of India In: Balasubramanian D, Rao NA, eds. The Indian Human Heritage. Hyderabad, India: Universities Press; 1998. p. :95-9.
- [Google Scholar]
- The racial affinities of the people of India In: Census of India, 1931, Part III-Ethnographical. Simla, India: Government of India Press; 1935.
- [Google Scholar]
- Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331-4.
- [CrossRef] [Google Scholar]
- Upper Paleolithic Siberian genome reveals dual ancestry of native Americans. Nature. 2014;505:87-91.
- [CrossRef] [PubMed] [Google Scholar]
- Reconstructing Indian population history. Nature. 2009;461:489-94.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93:422-38.
- [CrossRef] [PubMed] [Google Scholar]
- Examining the Farming/Language Dispersal Hypothesis Cambridge, England: McDonald Institute for Archaeological Research; 2003. p. :191-213.
- [Google Scholar]
- Petraglia MD, Allchin B, eds. The Evolution and History of Human Population in South Asia. Dordrecht, Netherlands: Springer; 2007. p. :393-443.
- [CrossRef]
- Genetics Proves Indian Population Mixture Massachusetts: Harvard Medical School; 2018.
- [Google Scholar]
- Who we are and how we Got Here: Ancient DNA and the New Science of the Human Past Oxford, United Kingdom: Oxford University Press; 2018. p. :1-368.
- [Google Scholar]
- A strong founder effect for two NLRP7 mutations in the Indian population: An intriguing observation. Clin Genet. 2009;76:292-5.
- [CrossRef] [PubMed] [Google Scholar]
- A founder variant in the South Asian population leads to a high prevalence of FANCL Fanconi anemia cases in India. Hum Mutat. 2019;41:122-8.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic reconstruction of the history of extant population of India reveals five distinct ancestral components and a complex structure. PNAS. 2016;113:1594-9.
- [CrossRef] [PubMed] [Google Scholar]
- Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic population. BMC Evol Biol. 2007;7:1-14.
- [CrossRef] [PubMed] [Google Scholar]
- YAP insertion signature in South Asia. Ann Hum Biol. 2009;34:582-6.
- [CrossRef] [PubMed] [Google Scholar]






